What do you know about Tuberculosis?
Background
Tuberculosis (TB) is a potentially serious infectious disease that mainly affects the lungs (called Pulmonary TB), but it can also affect almost any organ in the body. Once rare in developed countries, tuberculosis infections began increasing in 1985, partly because of the emergence of HIV, the virus that causes AIDS. India has the largest number of tuberculosis (TB) patients worldwide, accounting for over 25% of global cases.
Medications are available to treat TB and is administered as prescribed by the clinician. Depending on the medication(s) prescribed, the duration can be from four to nine months or more. Resistance to TB drugs is a formidable obstacle to effective TB care and prevention globally. Multidrug-resistant TB (MDR-TB) is multifactorial and fuelled by improper treatment of patients, poor management of supply and quality of drugs, and airborne transmission of bacteria in public places. As per the World Health Organisation (WHO) 2019 report, Drug resistant TB continues to be a public health threat. In 2018, there were about half a million new cases of rifampicin-resistant TB (of which 78% had multi drug resistant TB), and India stands one among the three high-burden countries accounting for 27% of the MDR/RR TB cases.
TB is transmitted through the air as tiny droplets when a person with TB sneezes, coughs, talks, laughs, or sings. Many a number of people are exposed to TB bacterium without becoming infected. Some can also become infected but may not develop active disease — this is called latent TB infection (LTBI). LTBI means that the immune system can contain TB infection and prevent active disease. Active TB disease happens when TB infection overwhelms the immune system, and the bacteria begin multiplying and cause disease.
1. Cough 2. Afternoon fever 3. Weight loss 4. Blood stained sputum 5. Night sweats
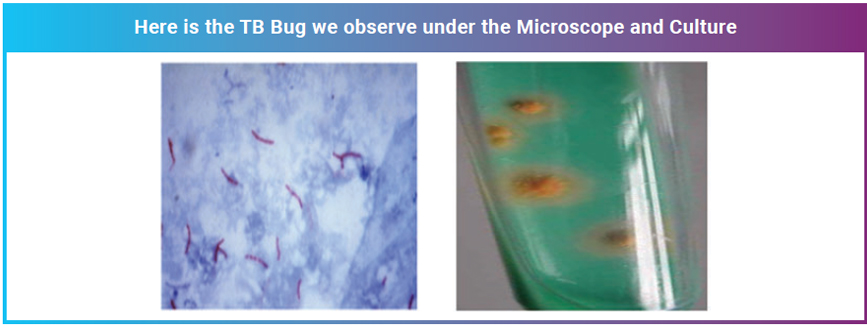
Mycobacterium tuberculosis (Mtb), the causative organism of tuberculosis, is a slow growing bacterium taking several weeks (4 to 6 weeks) for culture growth. This delay in diagnosis could worsen illness severity, prolong patient suffering, increase the risk of patient death, and facilitate the transmission of the disease (if smear-positive pulmonary TB) to close contacts
There are now 6 WHO-endorsed tests for detection of drug resistance: liquid culture, line-probe assays (LPA/Hains Test), the microscopically observed drug-susceptibility (MODS) assay, nitrate reductase assays (NRAs), colorimetric redox indicator (CRI) methods, and Xpert MTB/RIF (GeneXpert/CBNAAT test). Diagnosis of Mtb by culture remains the gold standard while Xpert MTB/RIF test and LPA, the two widely used molecular tests are replacing by their wide usage both by the public and the private sectors. Cultures take several weeks to diagnose Mtb varying from 6 days to 8 weeks. This is laborious and needs expert training. Xpert MTB/RIF could be placed in peripheral laboratories but only for identification of single drug (rifampicin) resistance with a reported sensitivity of 98% in smear positive cases and 70% in smear negative cases.
Line-probe assays could be placed at an intermediate level in the healthcare system but have low sensitivity on smear-negative sputum samples. Despite the advantage of these molecular tests on the timeline of diagnosis with direct specimens, the limitation of these are the detection of resistance to limited loci and does not eliminate the need for conventional culture and drug susceptibility testing (DST).
Whole-genome sequencing (WGS) has become a common technique for the investigation of pathogenic and environmental bacteria and has been used to address various aspects of tuberculosis. Culture-based WGS has been shown to reliably predict not only the drug resistance but also explain the differences in transmissibility between strains, or why some strains are more virulent than others or more prone to the development of multidrug resistance. In spite of the wide application of this technique, WGS has not yet become a predominant diagnostic in high-burden settings, due to the cost and time associated with culture growth.
Whole genome sequencing (WGS) offers the opportunity to screen not only the loci included in rapid molecular tests but also other known resistance-associated loci not screened by them. This enables the identification of new drug resistance-associated mutations that are not explained by currently available diagnostics at significantly shorter turnaround time. WGS performed directly on sputum samples so far has had limited success due to low Mtb load in sputum, resulting in low sequencing depth, which may under-identify drug resistance, with the presence of contaminating human and bacterial DNA, including non-tuberculous mycobacteria (NTMs) further adding analysis challenges.
Here is the TB Bug we observe under the Microscope and Culture For internal circulation in MedGenome only 19 To address the above challenges, we at MedGenome performed WGS on uncultured direct sputum samples to detect Mtb and identify drug resistance using a targeted enrichment-based method, followed by an improvised analysis technique to isolate Mtb-specific reads. The resistance variants predicted by this method were validated by comparing results to alternative technologies like Xpert, LPA, and culture-based phenotypic DST on the same samples. The Validated Test is the newly launched TB test - SPITSEQ – Whole Genome Sequencing of Mtb from Direct Sputum.
SPITSEQ, a Single test for diagnosis and drug resistance prediction, is a culture- free WGS method. This has 100% sensitivity and 98.4% to resistance variants profiled by line probe assay (LPA) and 97.7% accuracy with the phenotypic drug susceptibility tests for six anti-tuberculosis drugs using as little as 5ng of sputum DNA. This assay is a comprehensive drug panel revealing the mutations not only to the currently available anti TB drugs but also the newer drugs that are being validated.

From a cost perspective, WGS is more expensive than individual tests for smear, Gene Xpert, or LPA. However, WGS, as a standalone replacement for all these tests, along with culture-based DST, is more reasonably priced with an advantage of the lesser turnaround time. In light of the strong performance of this technique to predict drug-resistance compared to current tests and the greater information obtained from WGS, this method represents a possible tool for implementation in low-income, high-TB burden countries.
Soundararajan et al, Whole genome enrichment approach for rapid detection of Mycobacterium tuberculosis and drug resistance-associated mutations from direct sputum sequencing. Tuberculosis 121 (2020) 101915.