What is Imatinib Resistance Testing?

- Imatinib is a targeted tyrosine kinase inhibitor for patients suffering from Chronic Myeloid Leukemia (CML)
- Patients’ response to Imatinib has been significantly better and with fewer side effects than other available therapies
- But even in the best responders, some BCR-ABL1 positive cells usually remain.
- Patients who initially responded well to Imatinib have at times relapsed due to the development of resistance caused by changes in the ABL1 kinase domain. This affects Imatinib’s ability of binding to the active site

Prevalence

- CML accounts for 15% to 20% of all adult leukemias
- The diagnostic marker of CML is the Classical Philadelphia chromosome, t(9;22)(q34;q11)
- This results in the fusion of the N-terminal region of the BCR gene with the C-terminal kinase domain of the ABL1 gene. The fusion produces an active chimeric protein kinase responsible for leukemogenesis in CML.
- CML patients on Imatinib are monitored with quantitative RT-PCR and that patients with a log increase in quantitative BCR-ABL1 transcript levels are recommended to be assayed for the presence of Imatinib-resistance mutations
- Over 130 mutations in Imatinib-resistant patients are reported, of which frequent variations attributed to resistance include:
M237I M244V L248V G250E Q252H Y253F/H E255K/V D276G E279K V299L F311I/L/V T315I F317L M351T E355G F359V/C V379I L384M M388L H396R/P S417Y/T E450G E453K/V/D E459L/K/G F486S - The T315I mutation appears to confer resistance to multiple targeted tyrosine kinase inhibitors, while other mutations may be more responsive to other therapies
- Upon understanding the mutation status, the clinician intervenes by increasing the dosage of Imatinib depending on the exact mutation present. Or adding another kinase inhibitor may be effective in controlling BCR-ABL1 levels
- Determines the therapeutic strategy for each patient by identifying the mutation status of the ABL1 kinase domain by either:
- Increasing Imatinib dose
- Switching to second-generation inhibitors as Dasatinib, Nilotinib or Ponatinib
- Alternate treatment approaches
Why do you need the test?
Why MedGenome?

- MedGenome offers Imatinib Resistance testing by Next Generation Sequencing (NGS)
- Imatinib Resistance Test carried out by Next-generation Sequencing (NGS) technology has multiple advantages
- Sanger Sequencing could only detect mutations present above a 20% allele load
- RT-PCR has greater sensitivity, but limited by their short spectrum of mutation detection
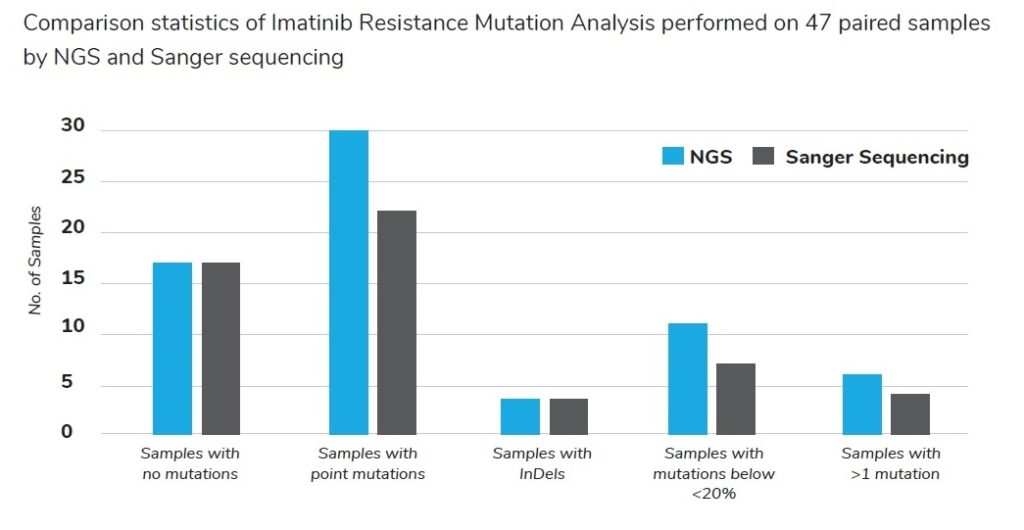
- Several NGS/Sanger comparison studies reported that Sanger sequencing had misclassified or underestimated kinase domain mutation status in up to 55% of samples, where mutations with 1-15% abundance are present
- NGS revealed emerging resistant mutants 2-11 months earlier when compared to Sanger sequencing
- Imatinib Resistance Mutation testing by NGS aids in:
- Full characterization of the spectrum of a minor (<20%) mutated variants
- Ability to follow the dynamics of resistant mutations over time
- Reconstruction of the clonal architecture of mutated populations in the case of multiple mutations occurring within the same amplicon.
Comparison between Imatinib Resistance Mutation Analysis by NGS, Sanger sequencing and RT-PCR
| Assay characteristics | NGS | Sanger sequencing | RT-PCR |
|---|---|---|---|
| Throughput | High | Low | Medium |
| The sensitivity of mutation detection | >1% | >20% | >5% |
| Ability to detect novel mutations | Yes | Yes | No |
| Identification of InDels | Yes | Yes | Yes |
| Distinguish between compound mutations | Yes | Yes | No |
| Identification of Polyclonal mutation | Yes | Yes | No |
| Quantification of mutation burden | Yes | Yes | No |
Validation Results:
| Sample type | Mutation type | Sensitivity | Specificity | Accuracy | Limit of detection |
|---|---|---|---|---|---|
| Peripheral blood RNA | SNV / Short-Indels | 100% | 100% | 100% | ≥1% |
Similar test
Get Genetic Counselling with MedGenome Genetic Experts
Please share your details with our genetic experts to answer your queries


 Enquire
Now
Enquire
Now